|
|
@@ -27,7 +27,7 @@ fextract <- function(x, y, keep = 1, top = TRUE) {
|
|
27
|
27
|
p <- import("pandas")
|
|
28
|
28
|
sns <- import("seaborn")
|
|
29
|
29
|
cbp <- as.character(p$Series(sns$color_palette("colorblind", as.integer(9))$as_hex()))
|
|
30
|
|
-aggdf <- p$read_pickle("../data/9-clusters.agg.pkl")
|
|
|
30
|
+aggdf <- p$read_pickle("../data/9-clusters-1617.agg.pkl")
|
|
31
|
31
|
# aggdf <- as.data.frame(aggdf)
|
|
32
|
32
|
aggdf$cluster <- factor(aggdf$cluster)
|
|
33
|
33
|
str(aggdf)
|
|
|
@@ -37,11 +37,11 @@ ggplot(aggdf, aes(y = kwh_tot_mean, x = cluster)) + geom_boxplot()
|
|
37
|
37
|
|
|
38
|
38
|
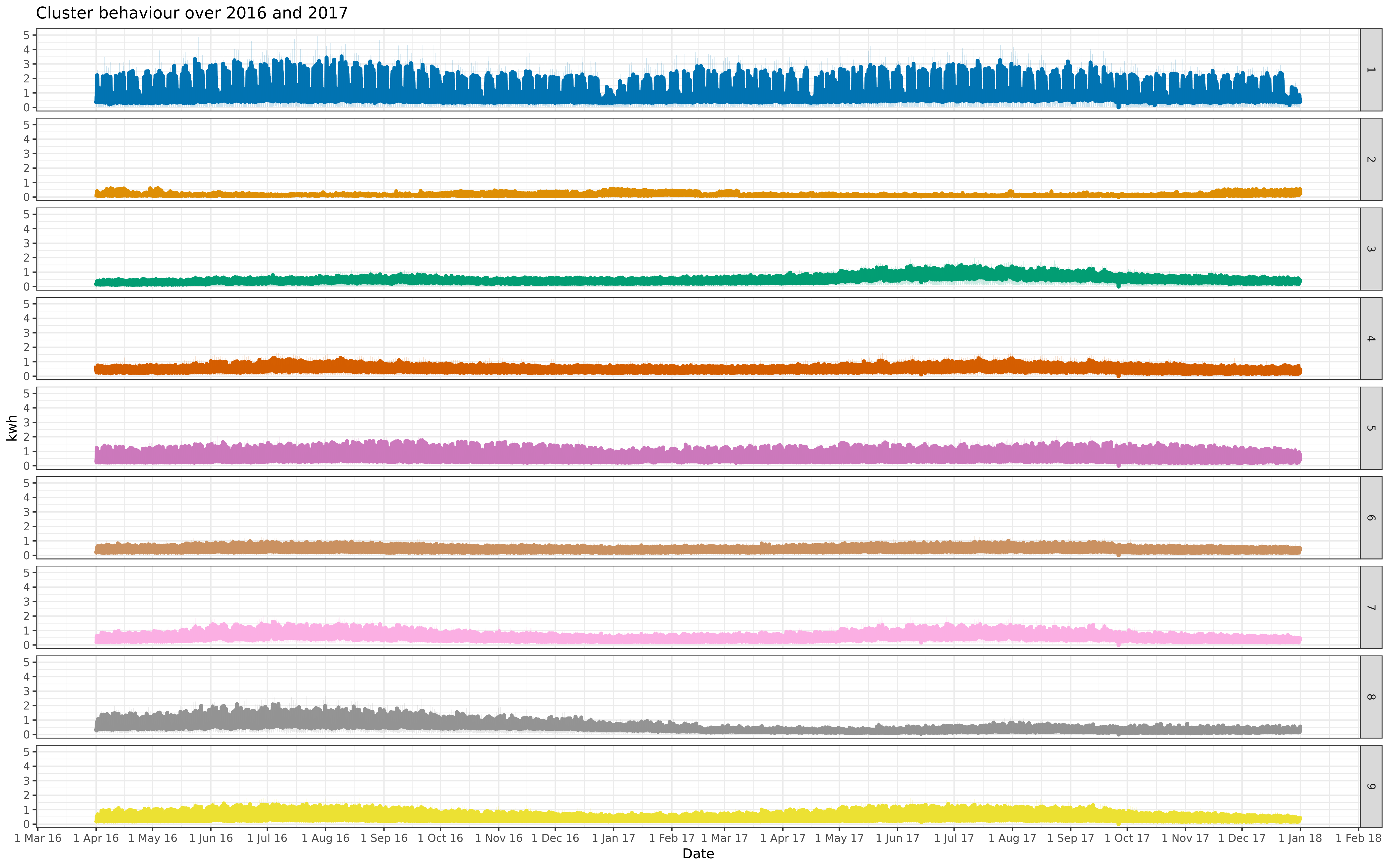
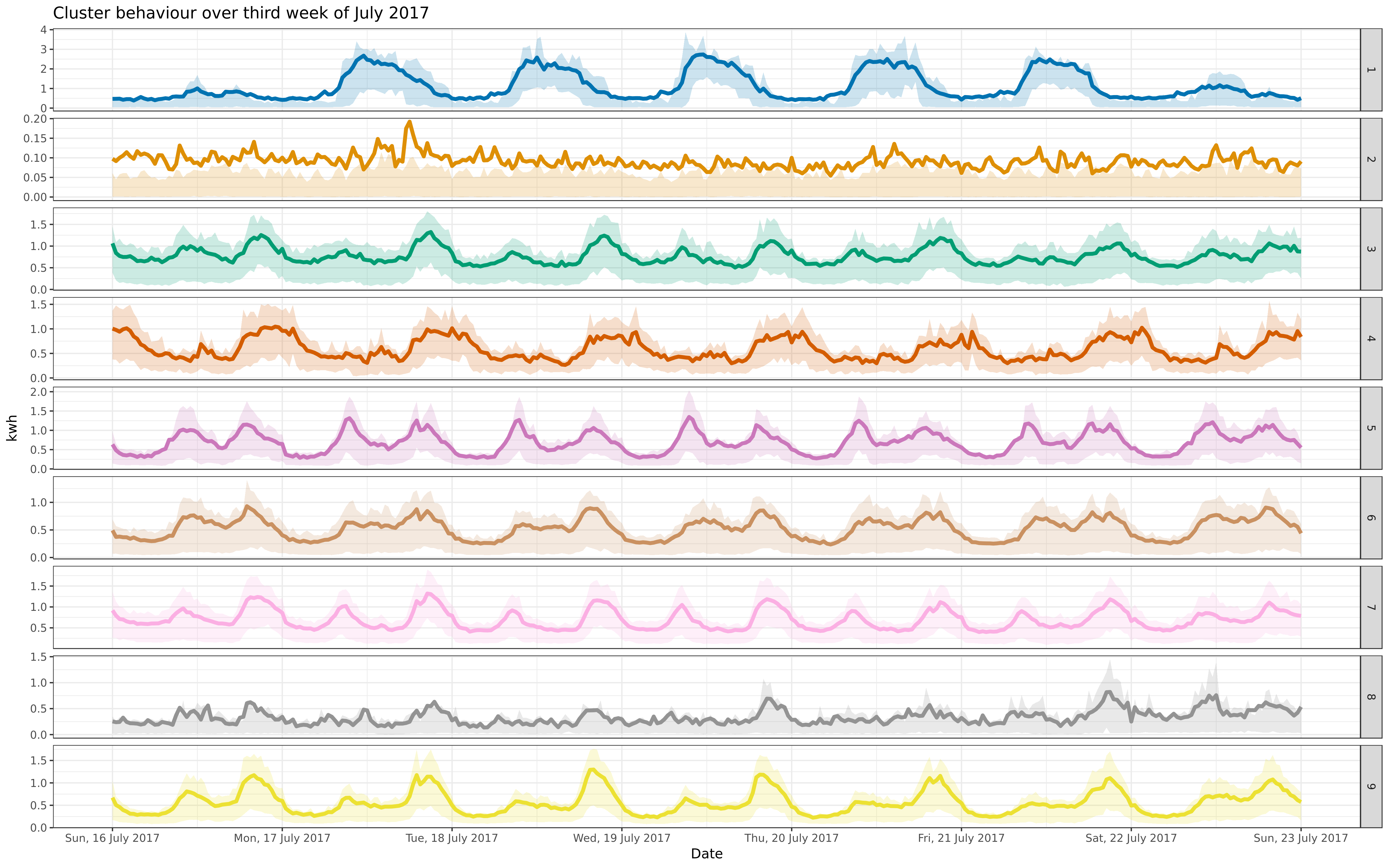
facall <- ggplot(aggdf, aes(x = read_time, y = kwh_tot_mean, color = cluster, fill = cluster)) +
|
|
39
|
39
|
geom_line(size = 1.5) + geom_ribbon(aes(ymin = kwh_tot_CI_low, ymax = kwh_tot_CI_high), alpha = 0.2, color = NA) +
|
|
40
|
|
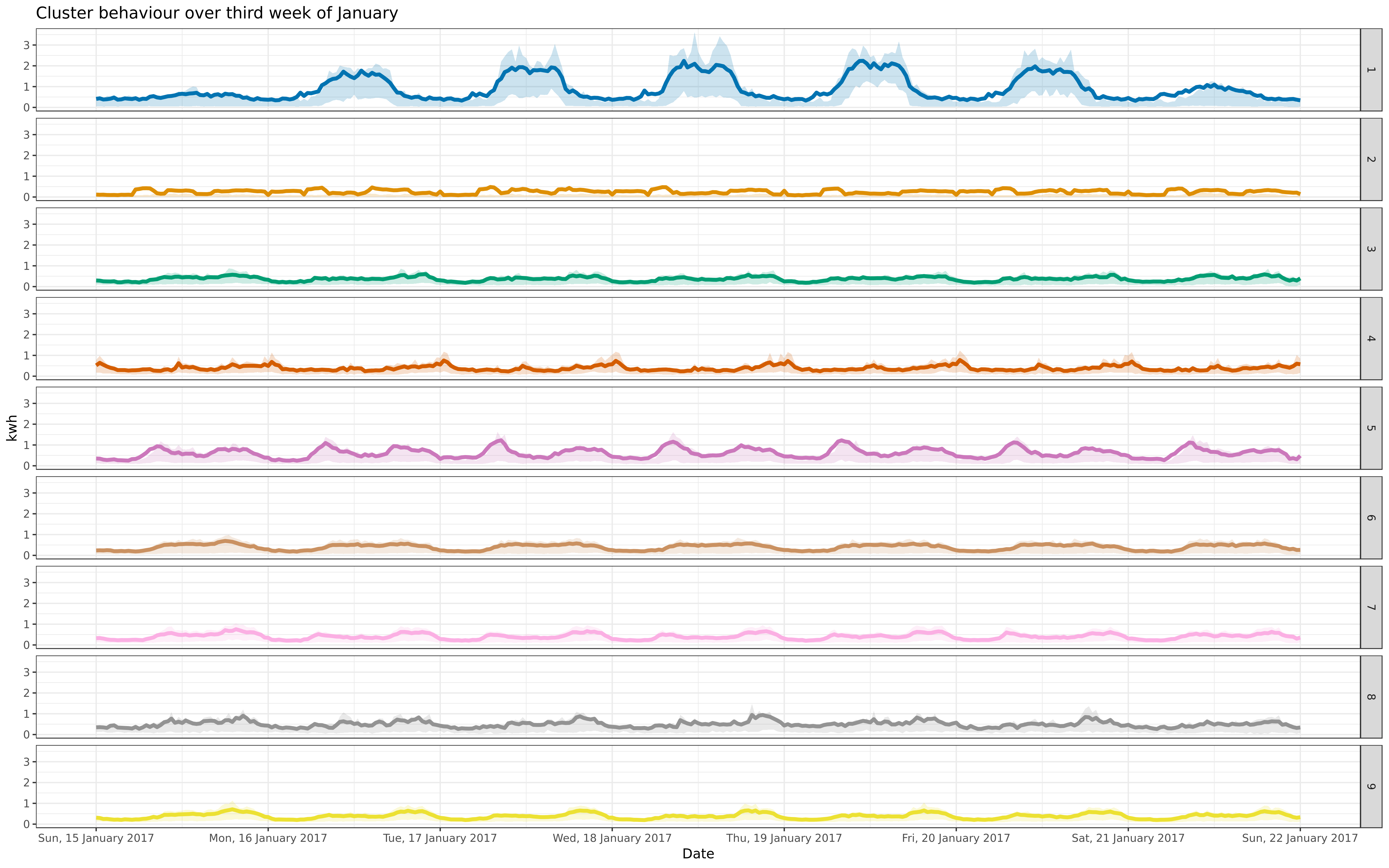
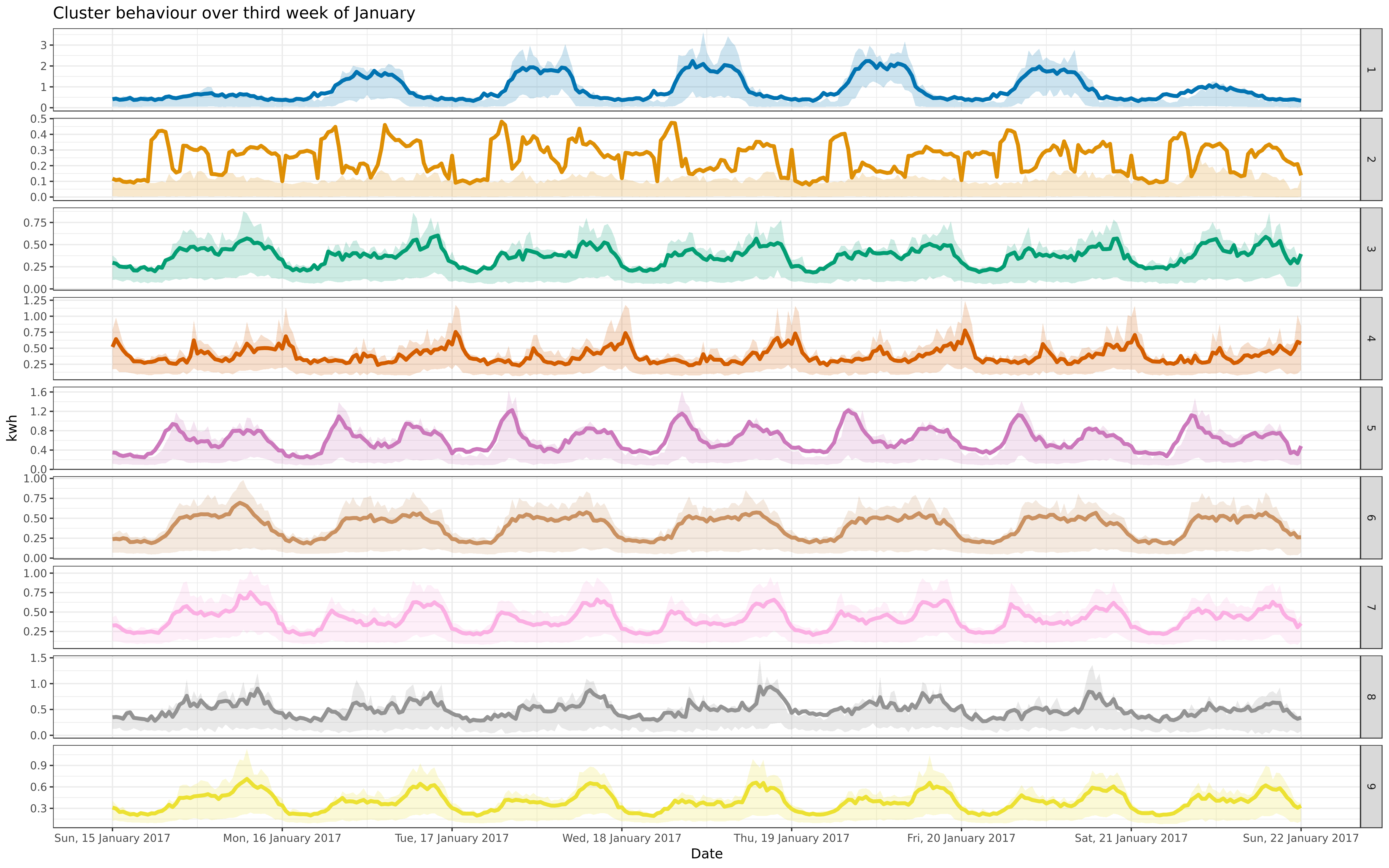
- labs(title = "Cluster behaviour over full year, 2017", x = "Date", y = "kwh") +
|
|
|
40
|
+ labs(title = "Cluster behaviour over 2016 and 2017", x = "Date", y = "kwh") +
|
|
41
|
41
|
scale_color_manual(values = cbp) +
|
|
42
|
42
|
scale_fill_manual(values = cbp) +
|
|
43
|
43
|
theme(legend.position = "none") +
|
|
44
|
|
- scale_x_datetime(date_breaks = "1 month", date_labels = "%-d %B")
|
|
|
44
|
+ scale_x_datetime(date_breaks = "1 month", date_labels = "%-d %b %y")
|
|
45
|
45
|
|
|
46
|
46
|
allcon <- facall + facet_grid(cluster ~ .)
|
|
47
|
47
|
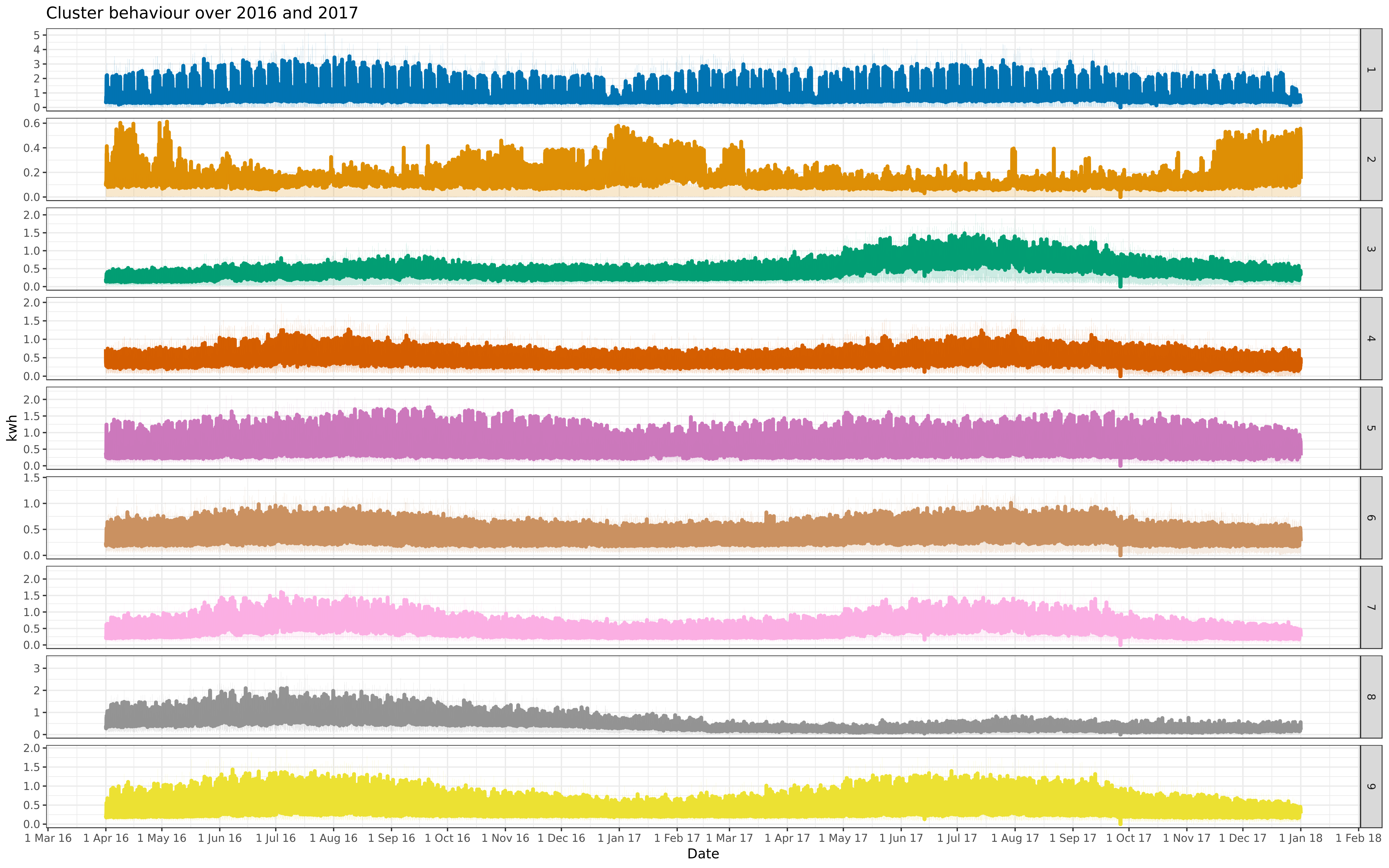
allfre <- facall + facet_grid(cluster ~ ., scales = "free")
|
|
|
@@ -101,16 +101,16 @@ facoct <- ggplot(midoct, aes(x = read_time, y = kwh_tot_mean, color = cluster, f
|
|
101
|
101
|
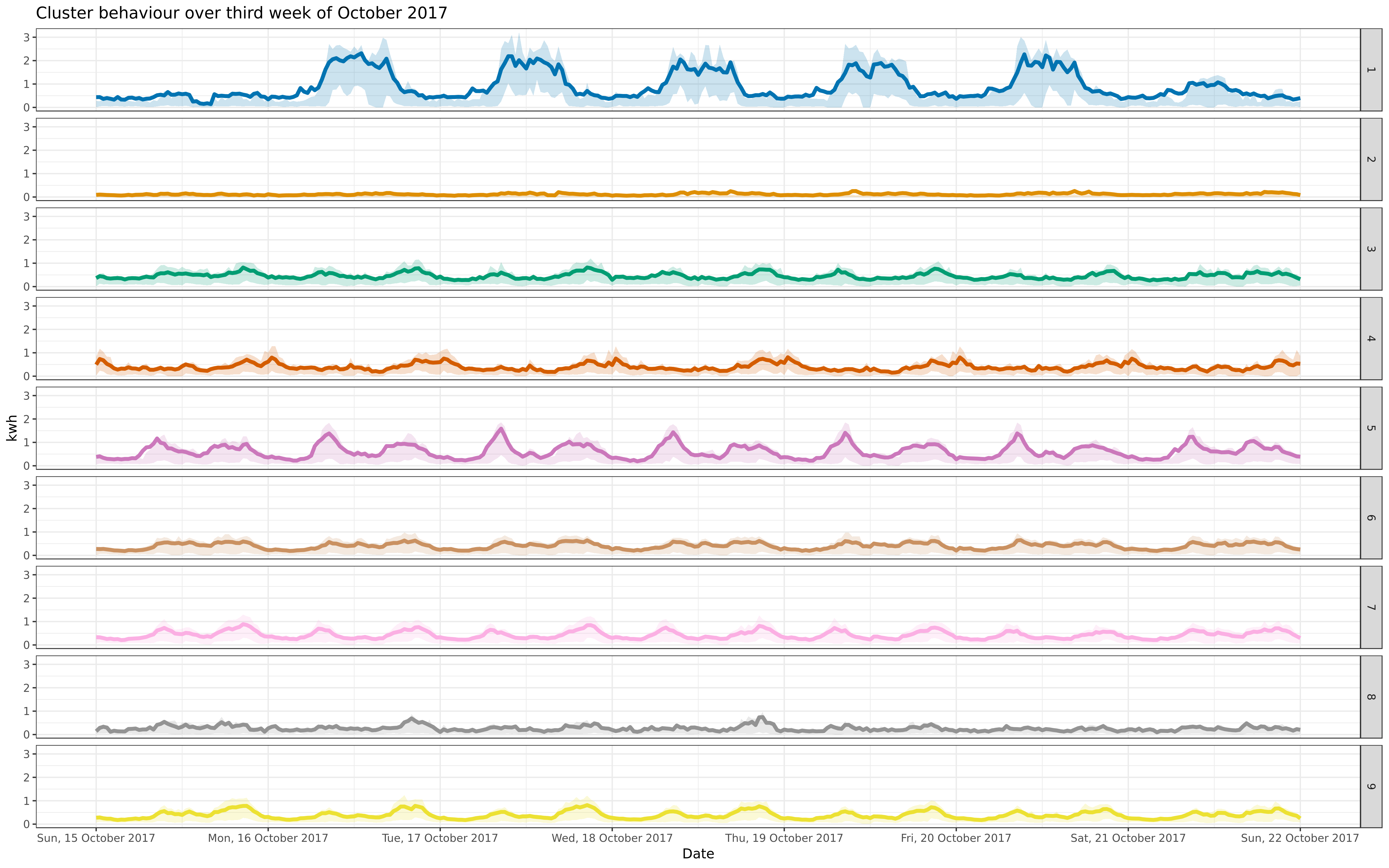
octcon <- facoct + facet_grid(cluster ~ .)
|
|
102
|
102
|
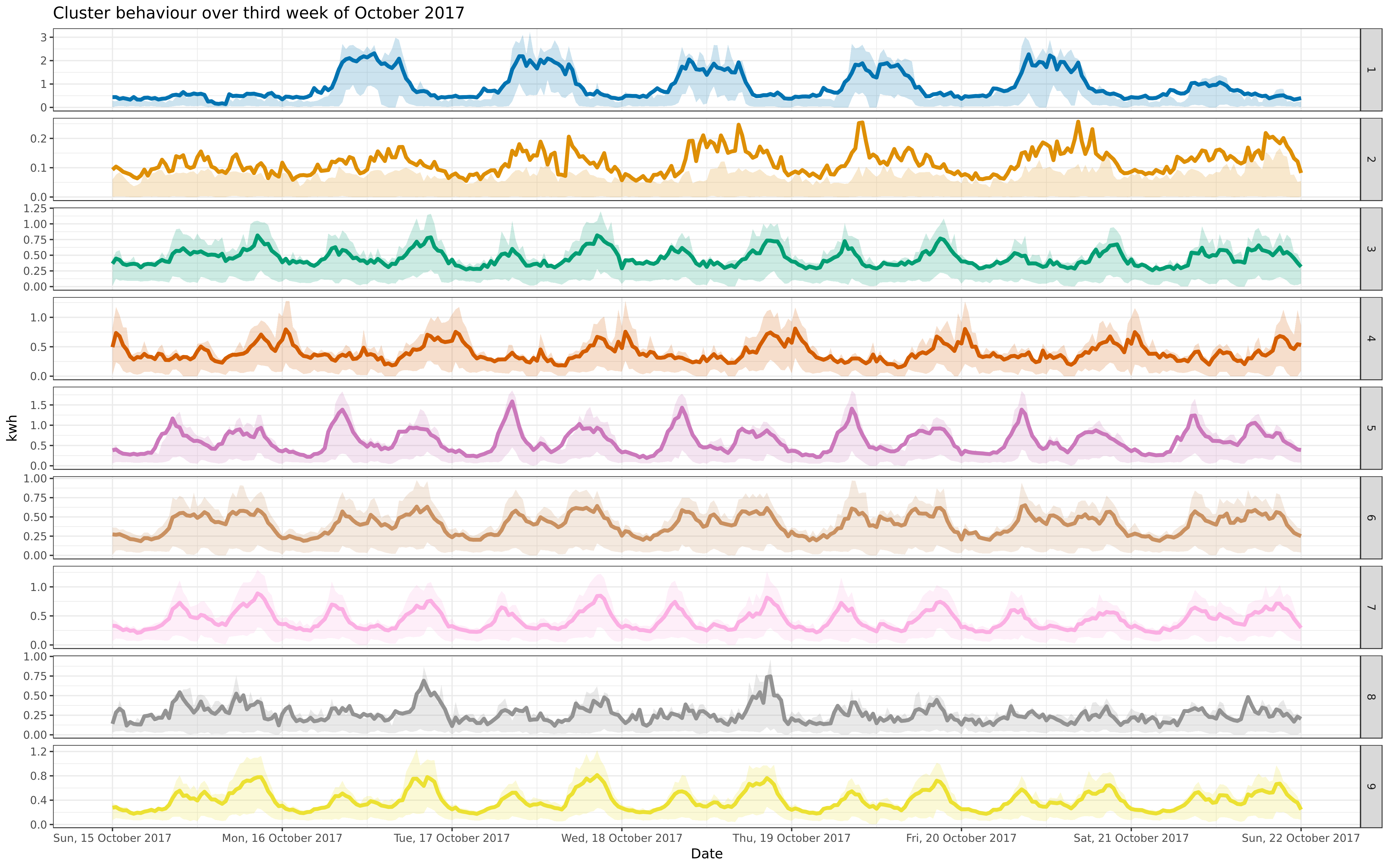
octfre <- facoct + facet_grid(cluster ~ ., scales = "free")
|
|
103
|
103
|
|
|
104
|
|
-ggsave("all-9-fix.png", allcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
105
|
|
-ggsave("all-9-fre.png", allfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
106
|
|
-ggsave("jan-9-fix.png", jancon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
107
|
|
-ggsave("jan-9-fre.png", janfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
108
|
|
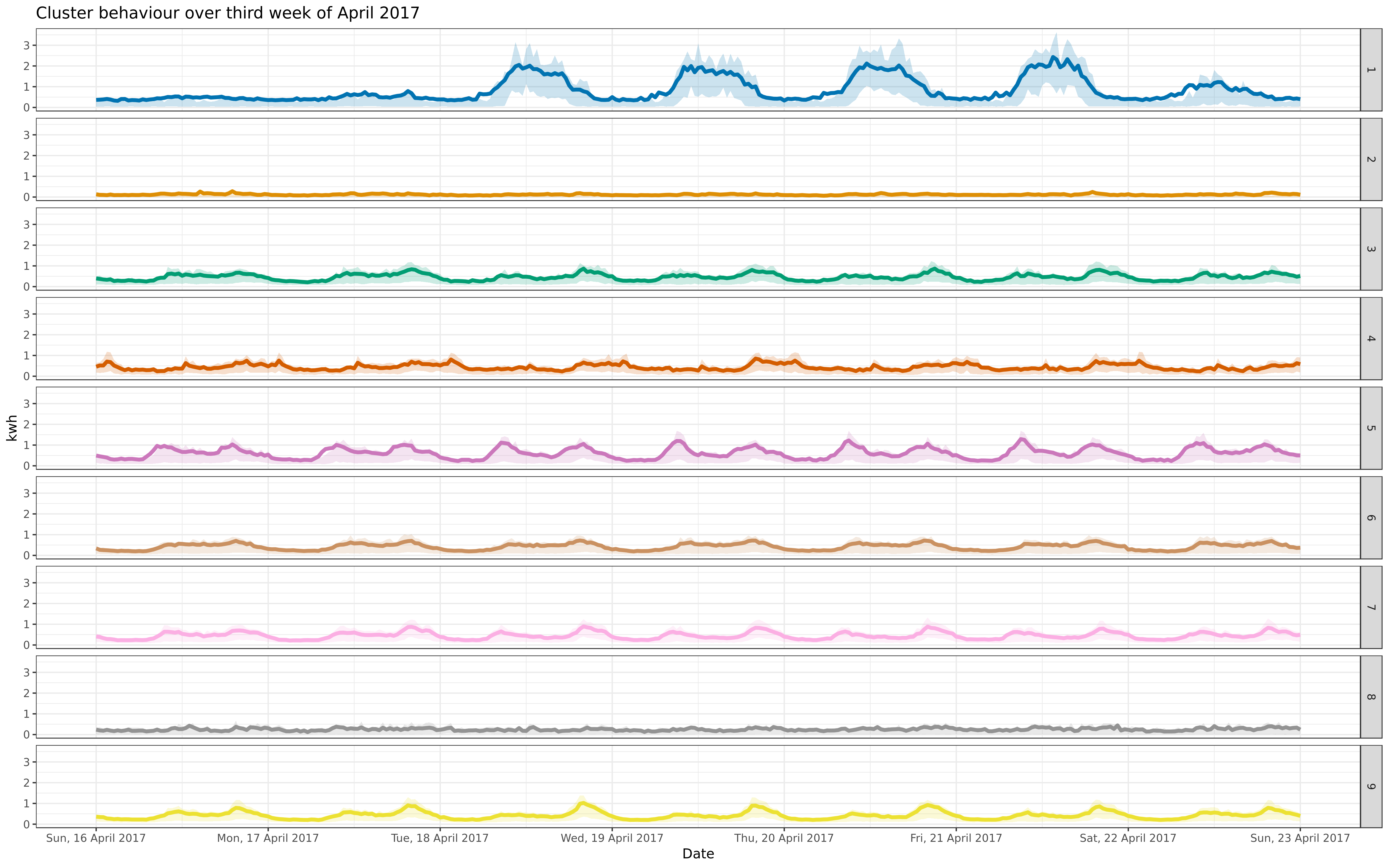
-ggsave("apr-9-fix.png", apcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
109
|
|
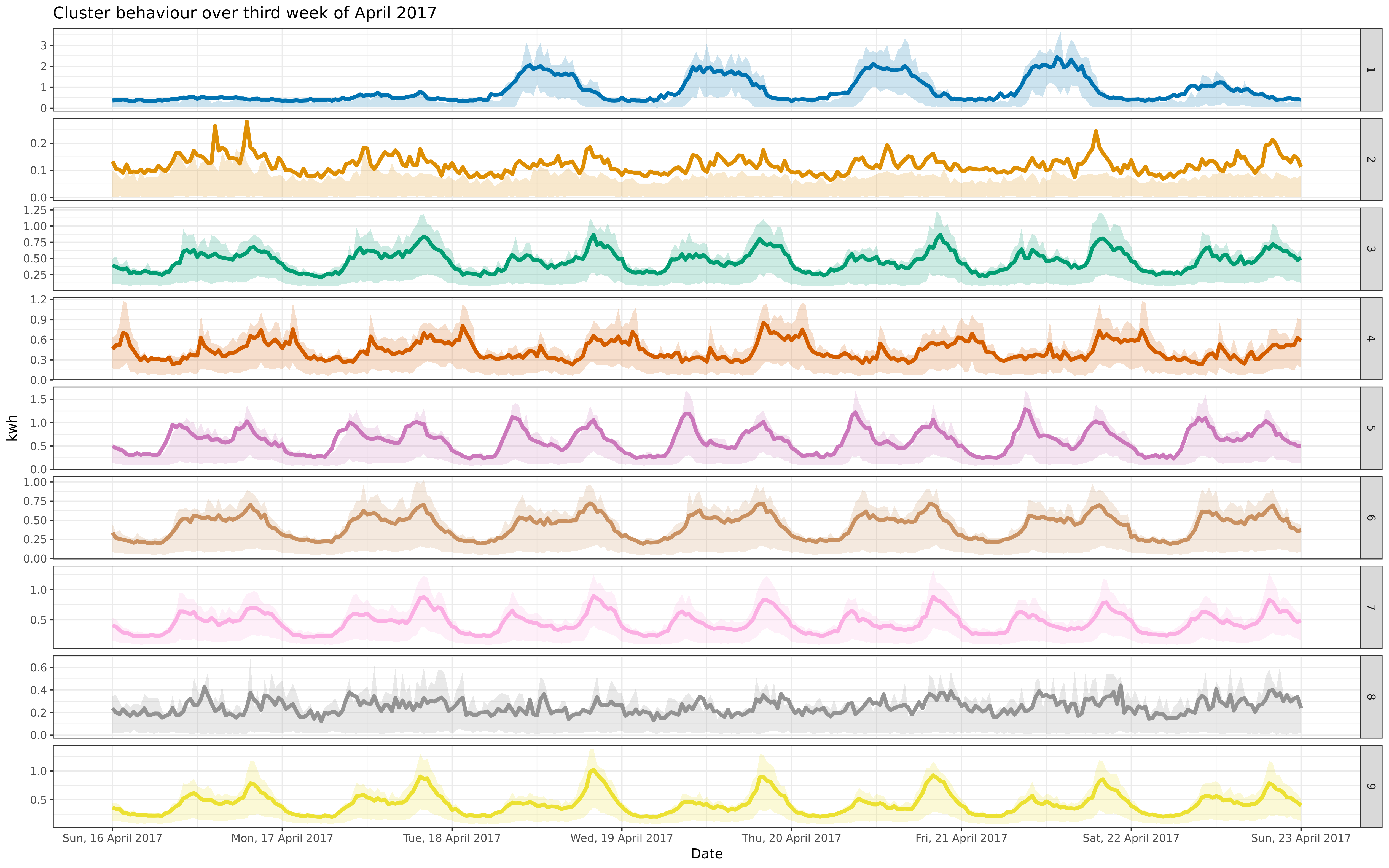
-ggsave("apr-9-fre.png", apfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
110
|
|
-ggsave("jul-9-fix.png", julcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
111
|
|
-ggsave("jul-9-fre.png", julfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
112
|
|
-ggsave("oct-9-fix.png", octcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
113
|
|
-ggsave("oct-9-fre.png", octfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
104
|
+ggsave("all-9-fix-1617.png", allcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
105
|
+ggsave("all-9-fre-1617.png", allfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
106
|
+ggsave("jan-9-fix-1617.png", jancon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
107
|
+ggsave("jan-9-fre-1617.png", janfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
108
|
+ggsave("apr-9-fix-1617.png", apcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
109
|
+ggsave("apr-9-fre-1617.png", apfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
110
|
+ggsave("jul-9-fix-1617.png", julcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
111
|
+ggsave("jul-9-fre-1617.png", julfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
112
|
+ggsave("oct-9-fix-1617.png", octcon, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
113
|
+ggsave("oct-9-fre-1617.png", octfre, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
114
|
114
|
|
|
115
|
115
|
|
|
116
|
116
|
# ----
|
|
|
@@ -142,8 +142,8 @@ wcorr <- ggplot(acfm, aes(x = day, y = acorr, color = cluster)) + geom_line(size
|
|
142
|
142
|
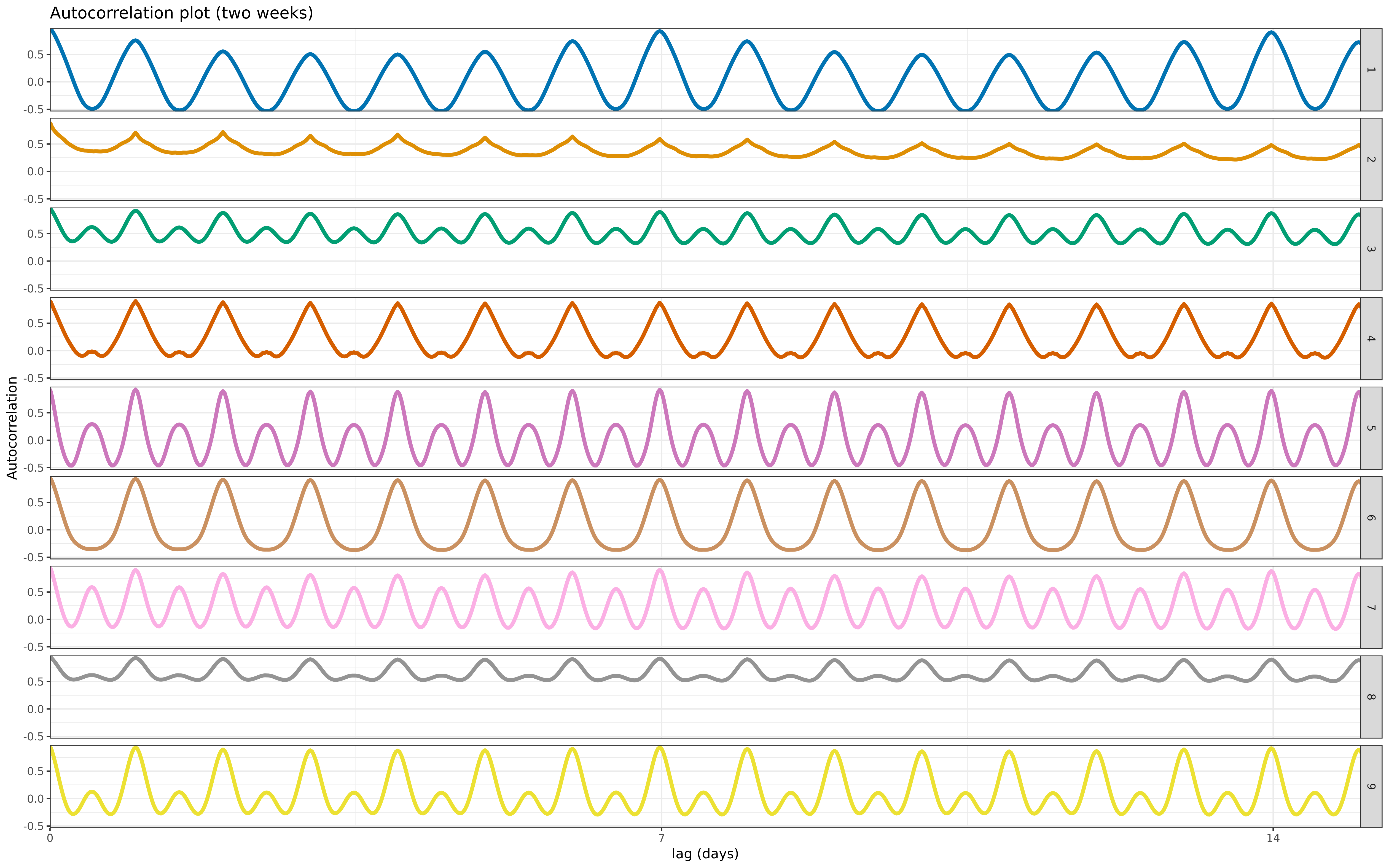
theme(legend.position = "none") + coord_cartesian(xlim = c(0, 15), expand = FALSE) +
|
|
143
|
143
|
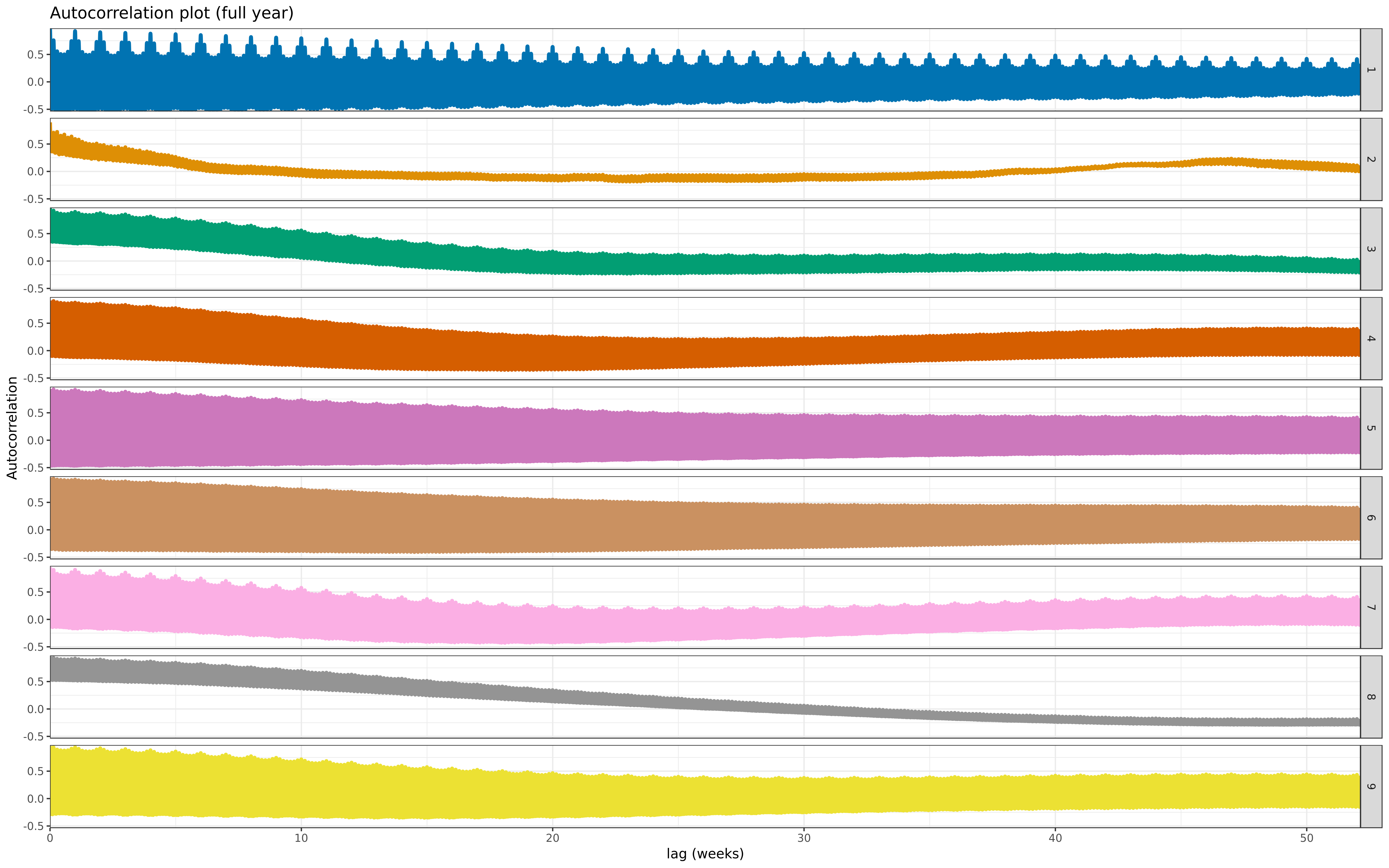
labs(title = "Autocorrelation plot (two weeks)", y = "Autocorrelation", x = "lag (days)")
|
|
144
|
144
|
|
|
145
|
|
-ggsave("full-autocorr.png", fcorr, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
146
|
|
-ggsave("week-autocorr.png", wcorr, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
145
|
+ggsave("full-autocorr-1617.png", fcorr, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
|
146
|
+ggsave("week-autocorr-1617.png", wcorr, path = "../img/", dpi = "retina", width = 40, height = 25, units = "cm")
|
|
147
|
147
|
|
|
148
|
148
|
perd <- bind_rows(perd)
|
|
149
|
149
|
|
|
|
@@ -160,9 +160,9 @@ ctbats <- tbats(ctsnp)
|
|
160
|
160
|
plot(forecast(ctbats, h = 48 * 7 * 4))
|
|
161
|
161
|
|
|
162
|
162
|
c9ts <- filter(aggdf, cluster == "9")$kwh_tot_mean
|
|
163
|
|
-ctsnp <- msts(c9ts, c(48, 48*7))
|
|
|
163
|
+ctsnp <- msts(c9ts, c(48, 48*7, 48*7*365.25))
|
|
164
|
164
|
ctbats <- tbats(ctsnp)
|
|
165
|
|
-plot(forecast(ctbats, h = 48 * 7 * 4))
|
|
|
165
|
+plot(forecast(ctbats, h = 48 * 7))
|
|
166
|
166
|
|
|
167
|
167
|
p <- periodogram(c1ts)
|
|
168
|
168
|
dd <- data.frame(freq = p$freq, spec = p$spec) %>% mutate(per = 1/freq)
|
|
|
@@ -172,11 +172,11 @@ c9ts <- filter(aggdf, cluster == "9")
|
|
172
|
172
|
|
|
173
|
173
|
ggplot(c9ts, aes(x = read_time, y = kwh_tot_mean)) + geom_line()
|
|
174
|
174
|
|
|
175
|
|
-nft <- fextract(c9ts$read_time, c9ts$kwh_tot_mean, keep = 10)
|
|
|
175
|
+nft <- fextract(c9ts$read_time, c9ts$kwh_tot_mean, keep = 15)
|
|
176
|
176
|
ggplot(nft, aes(x, y)) + geom_line() +
|
|
177
|
177
|
geom_line(aes(x, f), color = "blue") +
|
|
178
|
178
|
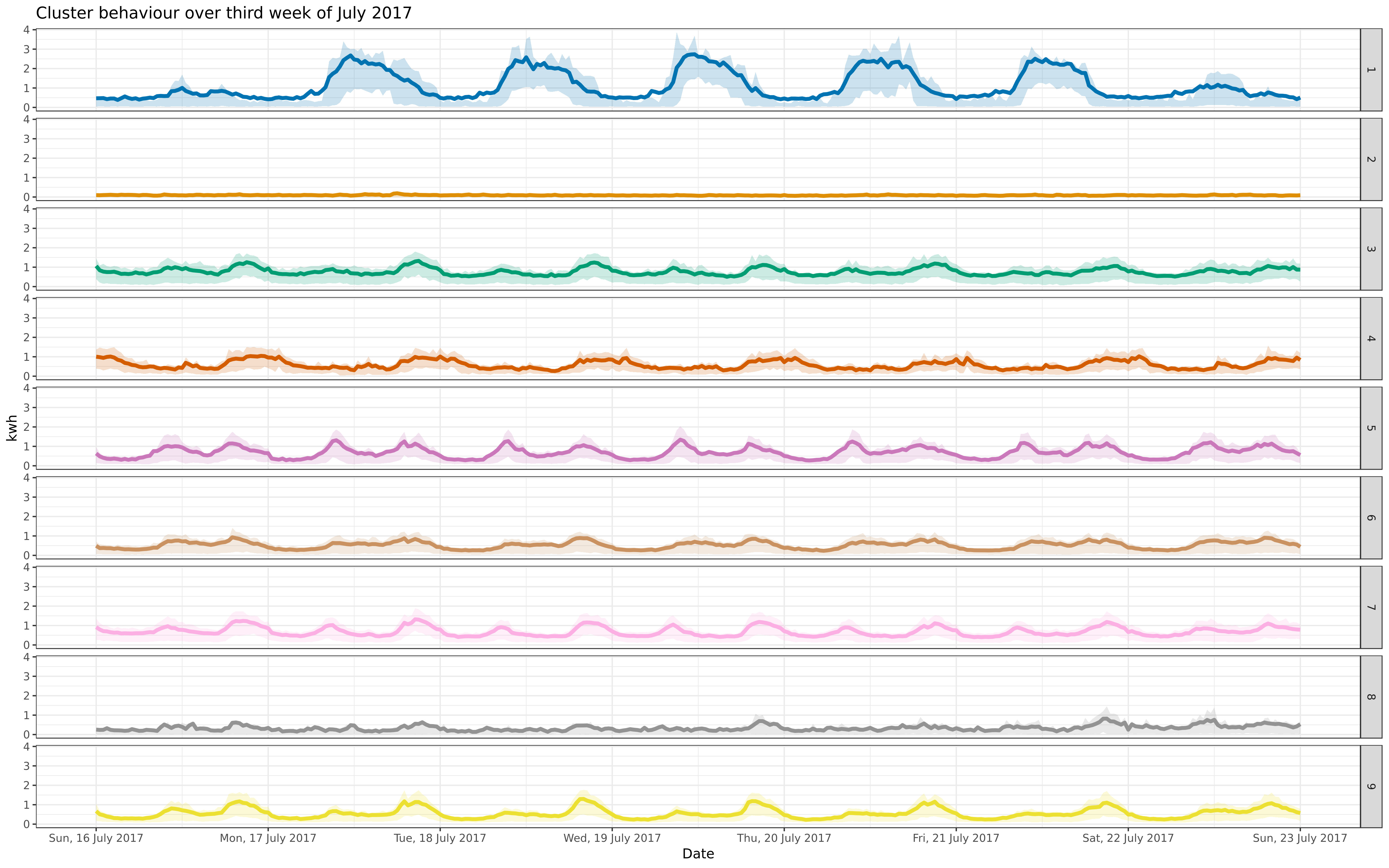
scale_x_datetime(date_breaks = "1 day", date_labels = "%a, %-d %B %Y") +
|
|
179
|
|
- coord_cartesian(xlim = c(as.POSIXct("2017-07-16", tz = "UTC"), as.POSIXct("2017-07-23", tz = "UTC")), expand = TRUE)
|
|
|
179
|
+ coord_cartesian(xlim = c(as.POSIXct("2016-07-16", tz = "UTC"), as.POSIXct("2016-07-23", tz = "UTC")), expand = TRUE)
|
|
180
|
180
|
|
|
181
|
181
|
clus <- "9"
|
|
182
|
182
|
kp <- 50
|